Genome Browsers: The Book of Life Isn’t Open Access
If I can access the human genome, does that make me a scientist? An analysis of scientific tools using the theory of cultural technique to understand how they structure social relations within a network.
Bioinformatics: A Technique Developed to Produce Knowledge
Science is a highly dynamic field of knowledge production. Every so often new technologies emerge that change the conditions necessary to produce knowledge and allow for great advances in our collective understanding. When the International Human Genome Sequencing Consortium (IHGS) published the first of the human reference genome in 2001, they fundamentally changed the field of biology. Through the comprehensive assemblage (map) they produced, scientists were able to demonstrate that the human genome contains only around 20,500 genes, which was much lower than the estimated 60,000 to 140,000 genes (NIH 2015). This meant that contrary to scientific belief, genes were not the source of organismal complexity. This realization forced them to consider that the key to “the Book of Life” might not lie in the genes themselves, but in what was referred to as “junk” DNA, the highly repetitive non-coding sequences of the human genome. In just a few years, the focus of genetic research shifted away from identifying genes to determining the elements that influence gene expression (the epigenome, the proteome, protein modifications, siRNA and other non-coding DNA elements).
As fundamental as this realization was for scientists, the completion of the Human Genome Project would not have been possible without computational analysis. In 1990, when scientists first attempted to sequence the genome in full, they were still using the laborious technique of cloning small sections of genetic code and inserting them into bacterial life forms. At the time, it was only possible to sequence short stretches of DNA using analogic methods. In order to sequence the three billion base pairs of the entire genome, it was necessary to use other techniques. While the project was publicly funded, it wasn’t until Celera, a private company, involved itself in the race for the human genome that computational analysis was used in sequencing techniques. Celera’s CEO, Craig Venter, had invented a new technique of DNA sequencing called “whole genome shotgun sequencing” that sequenced fragments of the entire genome simultaneously and used bioinformatics to reconstruct the sequence. While other sequencing technologies have since been developed, bioinformatics has remained central to the process of data assemblage and analysis.
Since the entire draft of the human genome was published in Nature over ten years ago, app developers have continued to produce platforms that enable users to browse the entire human genome free of charge. Genome browsers “intertextualize” assemblages of the human genome by displaying the different assemblages on tracks that can be configured by the end user. As a product of bioinformatics and the digital age of information sharing, genome browsers allow users to search the genome using gene location and map coordinates and to import their own sequences to compare their objects of study with previously identified genetic elements and their known functions. The use of genome browsers to analyze biological data has become a central practice of molecular biologists in identifying new genes and biological mechanisms.
The Cultural Technique of Genome Browsers: A Question of Accessibility
One way to consider how scientific tools shape social practice, according to the German theories of cultural technique, is to consider the history and development of the object as I have done. Genome browsers were developed in the wake of the Human Genome Project as a method of data analysis. Because of this, discourses of accessibility that emerged during genome mapping are applicable to processes that analyze the maps.
“Essentially, cultural techniques are conceived as operative chains that precede the media concepts they generate.” (Siegert 58)
Discourses of universal accessibility to the information produced by the publicly-funded project were strengthened by the UNESCO declaration in 1997 on the Human Genome and Human Rights. In a press release in 2001, the phrase “The information has been given away freely to the world—a vast and unique gift to celebrate the commonality of humankind” was used by the Sanger Institute, one of the 16 centres that formed the IHGS (Thomson 2001). In 1999, a scientist discussing the Human Genome Project within the context of a course wrote “To be valuable to society, genetic information must be available to all people in need of such information” (Boehm 1999).
In the same vein, the genome browser developed by the University of California-Santa Cruz (UCSC genome browser, 2002) is freely accessible online for educational purposes, and apps like GeneWall developed by Wobblebase, Inc. (2015) and Human Genome by Florence Haseltine (2012) are free to download. But is access the same as accessibility?
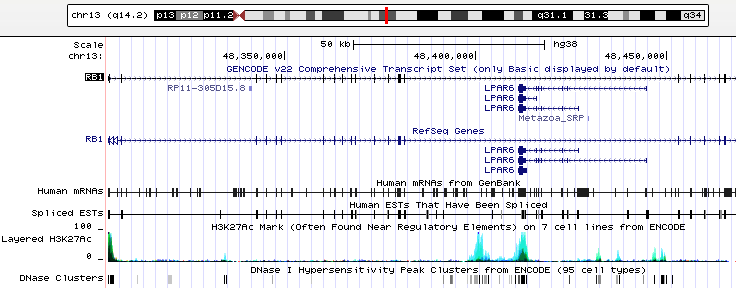
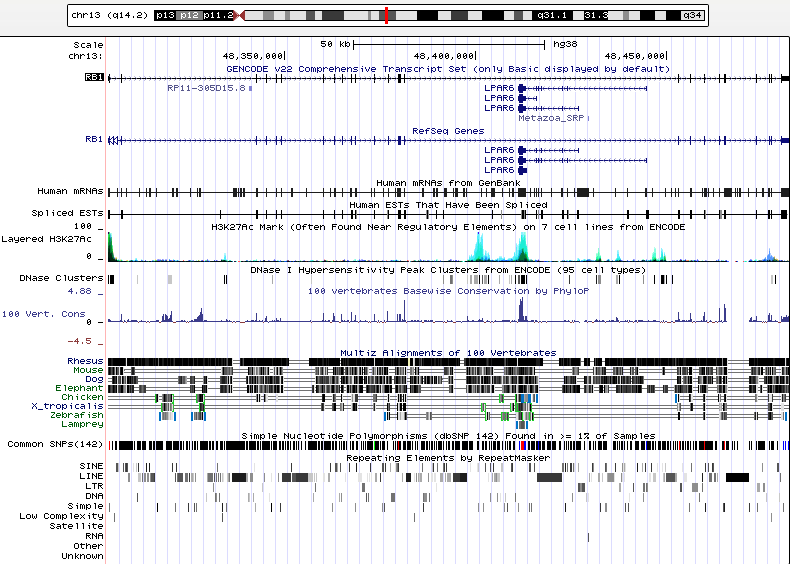
These genome browsers come in two flavors: research-oriented, like the UCSC genome browser, and user-friendly, like GeneWall. While these platforms are structurally similar, there are important differences in their infrastructure that determine their intended audience and user accessibility to database information.
The UCSC genome browser (pictured above) was developed much earlier than GeneWall’s and is the interface of choice in molecular biology labs. The interface’s native settings allows users to view a region on the genome that spans 26 assemblage tracks. The website allows sequence search and comparison (through tools BLAT) and allow users to navigate and customize the viewing of assembly sequences, mRNA transcripts and other organisms easily and quickly. However, navigating the database is complicated by the lack of explanation and presupposes users have a certain degree of biological literacy.
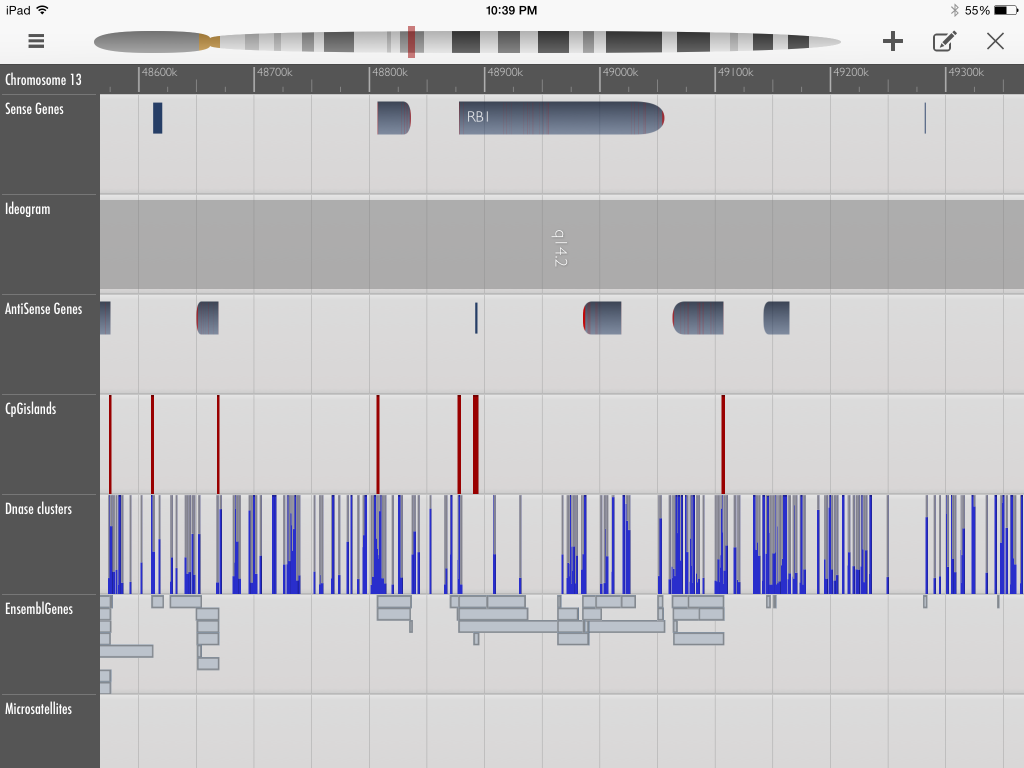
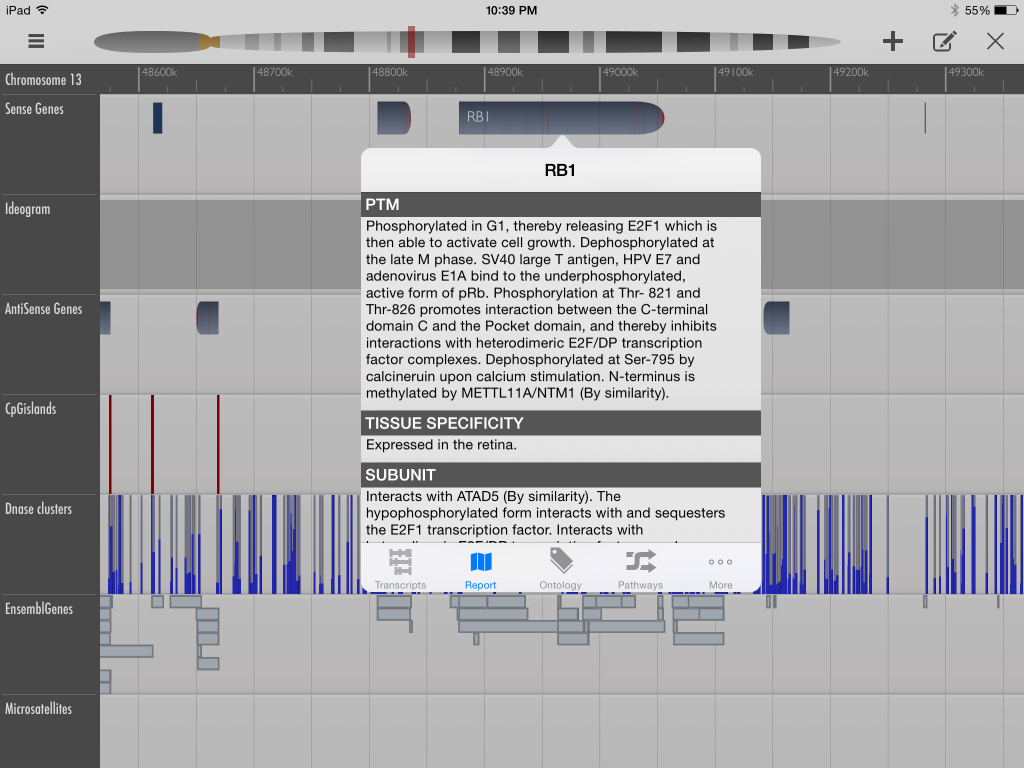
I have described GeneWall as more “user-friendly” because its interface, the iPad, allows users to navigate the assemblages using finger-swipe gestures. With a minimalist and uncluttered design, the descriptive elements for genes and other DNA elements are accessible by tapping on their mapped position. However, the system is in its infancy and is not as well connected to other tools like the UCSC browser. Another limitation of GeneWall is that its structure is centred around already a limited number of sequenced genes with determined functions, as opposed to displaying an array of mapped assemblages to allow users to infer the structure of the mapped region through juxtaposed elements. This user-friendly system also places limitations on the user’s potential production of scientific knowledge by focusing on the biological questions of the Human Genome Project era.
The Cultural Technique of Genome Browsers: A Tool to Limit User Access
These objects are structured to act as “gate-keepers” for scientific knowledge. By limiting the object’s users to “those with biological training” or limiting their databases to established forms of knowledge, developers of genome browsers are restricting the field that produces scientific knowledge. In this sense, the gatekeeping activity of these platforms confers authority to those with scientific training. In “The Growth of Medical Authority,” Paul Starr describes gatekeeping as the basis for professionalization: “The basis of modern professionalism has to be reconstructed around the claim to technical competence, gained through standardized training and evaluation” (475). This is made clear when you consider the access to content in MyGenome, developed by Illumina, a private biotech company: users are readily provided with an example genome, but Illumina will only release their personal sequence to the ordering physician. Despite all the rhetoric of the genome as human heritage that should be readily available, the truth is you can’t access your own genome until it’s been explained to you.
“Technologies are associated with habits and practices, sometimes crystallizing them and sometimes promoting them. They are structured by human practices so that they may in turn promote human practices.” (Sterne 376)
When I began reading media articles written by developers, I realized that they were defining access around infrastructures rather than the information they contain. They view genomics as a “niche product in large part used in and promoted by academia” (Kaganovich 2015) and instead are interested in developing “a wide variety of health and well-being apps and platforms that will be able to do things like connect variants to environmental, lifestyle, dietary, and activity-related factors, guiding both sick and healthy people towards a fundamental quality of life” (Menon 2015). Developers are interested in genomics that can be analyzed and marketed to the consumer without the need for the scientist. They see the future of genome browsers not as scientific tools but as gateways to personalized medicine.
Cultural Technique: A Method of Mapping the Actor-Network
Genome browsers were developed to allow users to access the human reference genome so that scientific knowledge would be advanced through data sharing. However, the limitations placed on users by the system infrastructures only allow users to interact with the information pre-determined by developers.
“When we speak of cultural techniques, therefore, we envisage a more or less complex actor network that comprises technological objects as well as the operative chains they are part of and that configure or constitute them.” (Siegert 58)
In this probe, in order to understand how genome browsers function as cultural techniques, I considered these tools within the context of their development to map the network of actors who interact with them. Actor-developers design these platforms with structures that control and limit the end-users’ possible interactions. This limiting aspect, that I have examined under the guise of “access” and “accessibility,” lends authority to a restricted group of end users who are able to obtain the platform’s information. This example illustrates the mechanisms of a cycle of knowledge production, within which objects are shaped by agent-developers to perform certain cultural actions and in turn shape agent-users by limiting access to information.
Works Cited
Boehm, David. “Applications and Issues of the Human Genome Project.” Plsc 431/631 Intermediate Genetics, ndsu.edu. web. 1999.
Kaganovich, Mark. “Genomics Needs a Killer App.” Crunch Network, techcrunch.com. web. March 27, 2015.
Menon, Prakash. “Coming Soon: An API for the human genome.” Health, venturebeat.com. web. June 27, 2015.
NIH. “An Overview of the Human Genome Project.” National Human Genome Research Institute. web. June 29, 2015.
Siegert, Bernhard. “Cultural Techniques: Or the End of the Intellectual Postwar Era in German Media Theory.” Theory, Culture & Society 30(6) 2013: 48-65.
Starr, Paul. “The Growth of Medical Authority.” Perspectives in Medical Sociology, 4th edition. Ed. Brown P. Long Grove, IL: Waveland Press, pp. 475-482.
Sterne, Jonathan. “Bourdieu, Technique and Technology.” Cultural Studies 17(3/4) 2003: 367-389.
Thomson, Mark. “The first draft of the Book of Humankind has been read.” Wellcome Trust Sanger Institute, sanger.ac.uk. web. June 26, 2001.

Over a summer a couple of years ago, I watched this fantastic lecture series by Stanford Professor Robert Sapolsky; it’s basically 25 videos of his course, “Human Behavioral Biology”, and if you’re like me and didn’t get quite enough science in your education, I’d recommend checking them out (even if you did, they’re worth watching for the way he helps you both to connect with and be skeptical of new theories and research). They really caught me up to speed and made it possible to read more widely with understanding on genes, genetics and brain science. (The first lecture will lead you to all the rest)
Here’s a short extract specifically describing how non-coding DNA elements work
For another angle on the collection of genetic information, consider how complicit these services (23andMe, ancestry.com) are with law enforcement.
“Both [these sites] stipulate in their privacy policies that they will turn information over to law enforcement if served with a court order”.
It’s not explicit that these sites have given over private genetic information to law enforcement, but why wouldn’t they?
Both sites also have the option to remove your genetic information from the database. However, their data-storage practices seem completely opaque – there’s no way to verify that they actually do it.
“[T]he fact that your signing up for 23andMe or Ancestry.com means that you and all of your current and future family members could become genetic criminal suspects is not something most users probably have in mind when trying to find out where their ancestors came from”.
further reading:
http://fusion.net/story/215204/law-enforcement-agencies-are-asking-ancestry-com-and-23andme-for-their-customers-dna/